Data.table: fread with large csv (44 GB) takes a lot of RAM in latest data.table dev version
Hi,
Hardware and software:
Server: Dell R930 4-Intel Xeon E7-8870 v3 2.1GHz,45M Cache,9.6GT/s QPI,Turbo,HT,18C/36T and 1TB in RAM
OS:Redhat 7.1
R-version: 3.3.2
data.table version: 1.10.5 built 2017-03-21
I'm loading a csv file (44 GB, 872505 rows x 12785 cols). It loads very fast, in 1.30 minutes using 144 cores (72 cores from the 4 processors with hyperthreading enabled to make it 144 cores box).
The main issue is that when the DT is loaded the amount of memory on-use increases significantly in relation to the size of the csv file. In this case the 44 GB csv (saved with fwrite, saved with saveRDS and compress=FALSE creates a file of 84GB) is using ~ 356 GB of RAM.
Here is the output using "verbose=TRUE"
_Allocating 12785 column slots (12785 - 0 dropped)
madvise sequential: ok
Reading data with 1440 jump points and 144 threads
Read 95.7% of 858881 estimated rows
Read 872505 rows x 12785 columns from 43.772GB file in 1 mins 33.736 secs of wall clock time (affected by other apps running)
0.000s ( 0%) Memory map
0.070s ( 0%) sep, ncol and header detection
26.227s ( 28%) Column type detection using 34832 sample rows from 1440 jump points
0.614s ( 1%) Allocation of 3683116 rows x 12785 cols (350.838GB) in RAM
0.000s ( 0%) madvise sequential
66.825s ( 71%) Reading data
93.736s Total_
It is showing a similar issue that sometimes arises when working with the parallel package, where one rsession is launched per core when using functions like "mclapply". See the Rsessions created/listed in this screenshot:

if I do "rm(DT)" RAM goes back to the initial state and the "Rsessions" get removed.
Already tried e.g. "setDTthreads(20)" and still using same amount of RAM.
By the way, if the file is loaded with the non-parallel version of "fread", the memory allocation only gets up to ~106 GB.
Guillermo
All 30 comments
This is the output from the non-parallel fread implementation (data.table 1.10.5 IN DEVELOPMENT built 2017-02-09)

And I double checked the amount of memory used and it just goes up to 84GB.
Guillermo
Yes you're right. Thanks for the great report. The estimated nrow looks about right (858,881 vs 872,505) but then the allocation is 4.2X bigger than that (3,683,116) and way off. I've improved the calculation and added more details to the verbose output. Hold off retesting for now though until a few more things have been done.
Ok please retest - should be fixed now.
I just installed data.table dev:
data.table 1.10.5 IN DEVELOPMENT built 2017-03-27 02:50:31 UTC
The first thing I got when I tried to read the same 44 GB file was this message:
DT <- fread('dt.daily.4km.csv')
Error: protect(): protection stack overflow
Then I just re-run the same command and started to work fine. However, this version is not using a multi-core mode. It is taking ~ 25 minutes to load, same as before you put the fread-parallel version.
Guillermo
I closed all r-sessions and re-run a test and I'm getting an error:
The guessed column type was insufficient for 34711745 values in 508 columns. The first in each column was printed to the console. Use colClasses to set these column classes manually.
See below, I get several messages regarding "guessed integer but contains <<0....>>
_Read 872505 rows x 12785 columns from 43.772GB file in 15:27.024 wall clock time (can be slowed down by any other open apps even if seemingly idle)
Column 171 ('D_19810618') guessed 'integer' but contains <<2.23000001907349>>
Column 347 ('D_19811211') guessed 'integer' but contains <<1.02999997138977>>
Column 348 ('D_19811212') guessed 'integer' but contains <<3.75>>_
Wow - your file is really testing the edge cases. Great. In future please run with verbose=TRUE and provide the full output. But with the information you provided in this case I can see what the problem is actually. There is a buffer created for each column for each thread (in this case, over 12,000 columns). Each one is separately PROTECTed currently. There is a way to avoid that - will do. The messages about the type guessing are correct. Do those 508 columns mean something to you and you agree they should be numeric? You can pass a range of columns to colClasses like this: colClasses=list("numeric"=11:518)
What field is this data from? Are you creating the file? It feels like it has been twisted to wide format where normal best practice is to write (and keep it in memory too) in long format. I'd normally expect to see the 508 columns names such as "D_19810618" as values in a column, not as columns themselves. Which is why I ask if you are creating the file and can you create it in long format. If not, suggest to whoever is creating the file that they can do it better. I guess that you are applying operations through columns perhaps using .SD and .SDcols. It's really much better in long format and keyby= the column holding the values like "D_19810618".
But I'll still try and make fread as good as it can be in dealing with any input -- even very wide files of over 12,000 columns.
I hope that other people are testing and finding no problems at all on their files!
_Do those 508 columns mean something to you and you agree they should be numeric? You can pass a range of columns to colClasses like this: colClasses=list("numeric"=11:518)_
This table contains time series by row. IDx,IDy, Time1_value, Time2_value, Time3_value... and all the TimeN_value columns contain only numeric values. If I use colClasses, I'm going to need to do it for 12783: list("numeric"=2:12783). I'll try this.
_What field is this data from?_
Geospatial data. I'm doing euclidean-distance searches within DT using IDx & IDy. I'm guessing that the more rows the slower the searches, right?
Right now is pretty fast (wide format). I have a map, where a user can click on an area and then a time series is generated within a csv file from the closest location (with data available) to the given click. I'll implement it with a long format instead.
I'll get back with some results.
Ok good. You don't need list("numeric"=2:12783) iiuc because it only needs help with 508 columns. Oh - I see - the 508 are scattered through the columns I guess then (they aren't a contiguous set of columns)?
No - data.table is almost never faster when it's wide! Long is almost always faster and more convenient. Have you seen and have you tried roll="nearest"? How are you doing it now? Please show the code so we can understand. Almost certainly long format is better but we may need some enhancements for 2D nearest. Please show the timings as well. When you say "pretty fast" it turns out that people have wildly different ideas about what "pretty fast" is.
melt'ing this table reach the 2^31 limit. I'm getting the error: "negative length vectors are not allowed".
I'll get back to the source to see if I can generate it in the long-format.
# Read Data
DT <- fread('dt.daily.4km.csv', showProgress = FALSE)
# Add two columns with truncated values of x and y (these are geog. coords.)
DT[,y_tr:=trunc(y)]
DT[,x_tr:=trunc(x)]
# For using on plotting (x-axis values)
xaxis<-seq.Date(as.Date("1981-01-01"),as.Date("2015-12-31"), "day")
# subset by truncated coordinates to avoid full-table search. Now searches
# will happen in a smaller subset
DT2 <- DT[y_tr==trunc(y_clicked) & x_tr==trunc(x_clicked),]
# Add distance from each point in the data.table to the provided location, "gdist" is from
# Imap package for euclidean distance.
DT2[,DIST:=gdist(lat.1 = DT2$y,
lon.1 = DT2$x,
lat.2 = y_clicked,
lon.2 = x_clicked, units="miles")]
# Get the minimum distance
minDist <- min(DT2[,DIST])
# Get the y-axis values
yt <- transpose(DT2[DIST==minDist,3:(ncol(DT2)-3)])$V1`
# Ready to plot xaxis vs yt
...
...
I don't have the application on a public server. Essentially, user can click on a map and then I capture those coordinates and perform the search above, get the time series, and create a plot.
Found and fixed another stack overflow for very large number of columns : https://github.com/Rdatatable/data.table/commit/d0469e670961dcdea115d433c0f2dce596d65906. Forgot to tag this issue number in the commit msg.
Oh. That's a point. 872505 rows * 12780 cols is 11 billion rows. So my suggestion to go long format won't work for you as that's > 2^31. Sorry - I should have spotted that. We'll just have to bite the bullet and go > 2^31 then. In the meantime let's stick with the wide format you have working and I'll nail that down.
Please try again. The memory usage should be back to normal and it should automatically reread the 508 out of 12,785 columns with out-of-sample type exceptions. To avoid the auto rerun time you can set colClasses.
If it's not fixed, please paste the full verbose output. Fingers crossed!
Ok...
A summary of results with the latest data.table dev: data.table 1.10.5 IN DEVELOPMENT built 2017-03-29 16:17:01 UTC
Four main points to mention:
- fread worked fine reading the file in the discussion.
- It took ~ 5.5 minutes to read vs. the previous version that took ~1.3 minutes.
- This version doesn't increase RAM allocation as the previous version I was testing.
- Cores seem to be "less active" (see below screenshot).
Some comments/questions:
1.
The percentage of usage in the cores is not as active as before, see screenshot:

In the previous version of fread, the cores' activity always was ~ 90-80%. In this version stayed around ~2-3 % each core as shown in the image above.
I don't understand why fread is doing bumps from integer to numeric: e.g.
Column 1489 ("D_19850126") bumped from 'integer' to 'numeric' due to <<0.949999988079071>> somewhere between row 6041 and row 24473
I double checked those rows and it seems ok. Why is detected as integer and need to be 'bumped' to 'numeric' if is already numeric (see summary for the rows suggested below)? or am I understanding this line wrongly? This happens for 508 lines. Are the NA's causing trouble?
summary(DT[6041:24473,.(D_19850126)])
D_19850126
Min. :0.750
1st Qu.:0.887
Median :0.945
Mean :0.966
3rd Qu.:1.045
Max. :1.210
NA's :18393
Below some verbose out of the test.
DT<-fread('dt.daily.4km.ver032917.csv', verbose=TRUE)
OUTPUT
Parameter na.strings == <<NA>>
None of the 0 na.strings are numeric (such as '-9999').Input contains no \n. Taking this to be a filename to open
File opened, filesize is 43.772296 GB.
Memory mapping ... ok
Detected eol as \n only (no \r afterwards), the UNIX and Mac standard.
Positioned on line 1 starting: <<x,y,D_19810101,D_19810102,D_19810103,D_19810104,D_19810105,D_19810106,D_19810107,D_19810108,D_19810109,D_19810110,D_19810111,D_19810112,D_19810113,
...
...
All the fields on line 1 are character fields. Treating as the column names.
Number of sampling jump points = 11 because 1281788 startSize * 10 NJUMPS * 2 = 25635760 <= -244636240 bytes from line 2 to eof
Type codes (jump 00) : 441111111111111111111111111111111111111111111111111111111111111111111111111111111111111111...1111111111 Quote rule 0
Type codes (jump 01) : 444422222222222222242444424444442222222222424444424442224424222244222222222242422222224422...4442244422 Quote rule 0
Type codes (jump 02) : 444422222222222222242444424444442422222242424444424442224424222244222222222242424222424424...4442444442 Quote rule 0
Type codes (jump 03) : 444422222222222224242444424444444422222244424444424442224424222244222222222244424442424424...4442444442 Quote rule 0
Type codes (jump 04) : 444444244422222224442444424444444444242244444444444444424444442244444222222244424442444444...4444444444 Quote rule 0
Type codes (jump 05) : 444444244422222224442444424444444444242244444444444444424444442244444222222244424442444444...4444444444 Quote rule 0
Type codes (jump 06) : 444444444422222224442444424444444444242244444444444444424444444444444244444444424442444444...4444444444 Quote rule 0
Type codes (jump 07) : 444444444422222224442444424444444444242244444444444444444444444444444244444444424444444444...4444444444 Quote rule 0
Type codes (jump 08) : 444444444442222224444444424444444444242244444444444444444444444444444444444444424444444444...4444444444 Quote rule 0
Type codes (jump 09) : 444444444442222424444444424444444444244444444444444444444444444444444444444444424444444444...4444444444 Quote rule 0
Type codes (jump 10) : 444444444442222424444444424444444444244444444444444444444444444444444444444444424444444444...4444444444 Quote rule 0
=====
Sampled 305 rows (handled \n inside quoted fields) at 11 jump points including middle and very end
Bytes from first data row on line 2 to the end of last row: 47000004016
Line length: mean=45578.20 sd=33428.37 min=12815 max=108497
Estimated nrow: 47000004016 / 45578.20 = 1031195
Initial alloc = 2062390 rows (1031195 + 100%) using bytes/max(mean-2*sd,min) clamped between [1.1*estn, 2.0*estn]
=====
Type codes (colClasses) : 444444444442222424444444424444444444244444444444444444444444444444444444444444424444444444...4444444444
Type codes (drop|select): 444444444442222424444444424444444444244444444444444444444444444444444444444444424444444444...4444444444
Allocating 12785 column slots (12785 - 0 dropped)
Reading 44928 chunks of 0.998MB (22 rows) using 144 threads
Read 872505 rows x 12785 columns from 43.772GB file in 05:26.908 wall clock time (can be slowed down by any other open apps even if seemingly idle)
Final type counts
0 : drop
0 : logical
0 : integer
0 : integer64
12785 : numeric
0 : character
Rereading 508 columns due to out-of-sample type exceptions.
Column 171 ("D_19810618") bumped from 'integer' to 'numeric' due to <<2.23000001907349>> somewhere between row 6041 and row 24473
Column 347 ("D_19811211") bumped from 'integer' to 'numeric' due to <<1.02999997138977>> somewhere between row 6041 and row 24473
Column 348 ("D_19811212") bumped from 'integer' to 'numeric' due to <<3.75>> somewhere between row 6041 and row 24473
Column 643 ("D_19821003") bumped from 'integer' to 'numeric' due to <<1.04999995231628>> somewhere between row 6041 and row 24473
Column 1066 ("D_19831130") bumped from 'integer' to 'numeric' due to <<1.46000003814697>> somewhere between row 6041 and row 24473
Column 1102 ("D_19840105") bumped from 'integer' to 'numeric' due to <<0.959999978542328>> somewhere between row 6041 and row 24473
Column 1124 ("D_19840127") bumped from 'integer' to 'numeric' due to <<0.620000004768372>> somewhere between row 6041 and row 24473
Column 1130 ("D_19840202") bumped from 'integer' to 'numeric' due to <<0.540000021457672>> somewhere between row 6041 and row 24473
Column 1489 ("D_19850126") bumped from 'integer' to 'numeric' due to <<0.949999988079071>> somewhere between row 6041 and row 24473
Column 1508 ("D_19850214") bumped from 'integer' to 'numeric' due to <<0.360000014305115>> somewhere between row 6041 and row 24473
...
...
Reread 872505 rows x 508 columns in 05:29.167
Read 872505 rows. Exactly what was estimated and allocated up front
Thread buffers were grown 0 times (if all 144 threads each grew once, this figure would be 144)
=============================
0.000s ( 0%) Memory map
0.093s ( 0%) sep, ncol and header detection
0.186s ( 0%) Column type detection using 305 sample rows from 44928 jump points
0.600s ( 0%) Allocation of 872505 rows x 12785 cols (192.552GB) plus 1.721GB of temporary buffers
326.029s ( 50%) Reading data
329.167s ( 50%) Rereading 508 columns due to out-of-sample type exceptions
656.075s Total
This last summary took a lot of time to finish it (~6 minutes). If we count this verbose summary the whole fread took ~ 11 minutes. It is re-reading those 508 columns, I can even see the message "Rereading 508 columns due to out-of-sample type exceptions" without using the verbose=TRUE.
Check it out without "verbose=TRUE"
ptm<-proc.time()
DT<-fread('dt.daily.4km.ver032917.csv')
Read 872505 rows x 12785 columns from 43.772GB file in 05:26.647 wall clock time (can be slowed down by any other open apps even if seemingly idle)
Rereading 508 columns due to out-of-sample type exceptions.
Reread 872505 rows x 508 columns in 05:30.276
proc.time() - ptm
user system elapsed
2113.100 85.919 657.870
fwrite test
It is working fine. Really fast. I thought that writing was slower than reading these days.
fwrite(DT,'dt.daily.4km.ver032917.csv', verbose=TRUE)
No list columns are present. Setting sep2='' otherwise quote='auto' would quote fields containing sep2.
maxLineLen=151187 from sample. Found in 1.890s
Writing column names ... done in 0.000s
Writing 872505 rows in 32315 batches of 27 rows (each buffer size 8MB, showProgress=1, nth=144) ...
done (actual nth=144, anyBufferGrown=no, maxBuffUsed=46%)
The "Reread" is taking quite of time. It is like reading the file twice.
Excellent! Thanks for all the info.
- Yes you're right it shouldn't be rereading. You've passed
colClasses=list("numeric"=1:12785)so the output line startingType code (colClasses)should be all value 4. Will fix and add missing test. - On 5.5m vs 1.3m, that's interesting. Each thread's buffer is currently 1MB. The idea being to be small to fit in cache. But in your case 1MB / 12785 cols = 82 bytes. So it's being 50x too cache inefficient with that choice. I guess, anyway. I'd have never thought of that without your testing. When it was working at 1.3m speed it didn't have that 1MB size in there.
- Why it has only sampled 305 lines is very strange too. It should have sampled 1,000. If that's fixed maybe it'll detect the 508 columns without you having to. The sample isn't taking any time (0.186s) so the sample size could be increased.
Can you paste the output of lscpu unix command please. This will tell us your cache sizes and I can think from there. I'll provide buffMB as a parameter to fread so you can see if it is that. I can come up with a better calculation if it is.
My mistake in one point of my previous post. The colClasses=list("numeric"=1:12785) is working fine. If I don't specify "colClasses" it does the "Rereading". Sorry about the confusion.
One thing that I noticed is that if I don't specify the "colClasses" the table is created with NA's and RAM shows as if the DT was loaded normal (~106MB in RAM).
Here is the lscpu output:
Architecture: x86_64
CPU op-mode(s): 32-bit, 64-bit
Byte Order: Little Endian
CPU(s): 144
On-line CPU(s) list: 0-143
Thread(s) per core: 2
Core(s) per socket: 18
Socket(s): 4
NUMA node(s): 4
Vendor ID: GenuineIntel
CPU family: 6
Model: 63
Model name: Intel(R) Xeon(R) CPU E7-8870 v3 @ 2.10GHz
Stepping: 4
CPU MHz: 2898.328
BogoMIPS: 4195.66
Virtualization: VT-x
L1d cache: 32K
L1i cache: 32K
L2 cache: 256K
L3 cache: 46080K
NUMA node0 CPU(s): 0,4,8,12,16,20,24,28,32,36,40,44,48,52,56,60,64,68,72,76,80,84,88,92,96,100,104,108,112,116,120,124,128,132,136,140
NUMA node1 CPU(s): 1,5,9,13,17,21,25,29,33,37,41,45,49,53,57,61,65,69,73,77,81,85,89,93,97,101,105,109,113,117,121,125,129,133,137,141
NUMA node2 CPU(s): 2,6,10,14,18,22,26,30,34,38,42,46,50,54,58,62,66,70,74,78,82,86,90,94,98,102,106,110,114,118,122,126,130,134,138,142
NUMA node3 CPU(s): 3,7,11,15,19,23,27,31,35,39,43,47,51,55,59,63,67,71,75,79,83,87,91,95,99,103,107,111,115,119,123,127,131,135,139,143
Ok got it. Thanks.
Reading your first comments again, would it make more sense if it said "bumping from integer to double" rather than "bumping from integer to numeric"?
What do you mean by 'created with NA's'? All the table is full of NAs, just the 508 columns?
What is 'DT was loaded normal (~106MB in RAM)'. It's a 44GB file so how can 106MB be normal?
Well, in this file all I have are real numbers and NA's. I don't have integer values in it. When do you throw that message, "bumping from datatypeA to datatypeB"?
That's exactly what I mean. In the current version of data.table that I installed, if I omit the colClasses it loads the DT but full of NA's.
DT[!is.na(D_19821001),]produces 0 records and if I load the table with colClasses and do the same filtering it actually shows the records.Well, this file is 47 GB as csv in disk, but once you load it into R, it takes more than double in RAM... Isn't related to the precision of the values, once is loaded the allocation of memory for real numbers cause that increase?
Maybe another typo then : 106MB should have been 106GB.
I'm not following the NA aspect. But a few fixes on the way and then try again fresh...
Ok - please try again. The guessing sample has increased to 10,000 (it'll be interesting to see how long that takes) and the buffer sizes now have a minimum imposed.
You may need to wait at least 30 mins for the drat package file to be promoted.
Sorry, it was "GB" instead of "MB". I didn't have enough coffee.
I'll get the update and test it.
Testing with latest data.table dev.: data.table 1.10.5 IN DEVELOPMENT built 2017-03-30 16:31:45 UTC :
Summary:
It is working fast (1.43 minutes for reading the csv) and memory allocation is working fine too, I guess, RAM allocation doesn't increase as it did before. A 44 GB csv on disk translates into a ~112 (+ 37 GB temp. buffers) GB in RAM once is loaded. Is this related to the data type of the values in the file?
Test 1:
Without using colClasses=list("numeric"=1:12785)
DT<-fread('dt.daily.4km.csv', verbose=TRUE)
Parameter na.strings == <<NA>>
None of the 1 na.strings are numeric (such as '-9999').
Input contains no \n. Taking this to be a filename to open
File opened, filesize is 43.772296 GB.
Memory mapping ... ok
Detected eol as \n only (no \r afterwards), the UNIX and Mac standard.
Positioned on line 1 starting: <<x,y,D_19810101,D_19810102,D_19810103,D_19810104,D_19810105,D_19810106,D_19810107,
D_19810108,D_19810109,D_19810110,D_19810111,D_19810112,
...
All the fields on line 1 are character fields. Treating as the column names.
Number of sampling jump points = 101 because 47000004016 bytes from row 1 to eof / (2 * 1281788 jump0size) == 18333
Type codes (jump 000) : 441111111111111111111111111111111111111111111111111111111111111111111111111111111111111111...1111111111 Quote rule 0
Type codes (jump 001) : 444222422222222224442444444444442222444444444444444444442444444444444222224444444444444444...4444444442 Quote rule 0
Type codes (jump 002) : 444222444444222244442444444444444422444444444444444444442444444444444222224444444444444444...4444444442 Quote rule 0
...
Type codes (jump 034) : 444444444444444444444444444444444444444444444444444444444444444444444444444444444444444444...4444444444 Quote rule 0
Type codes (jump 100) : 444444444444444444444444444444444444444444444444444444444444444444444444444444444444444444...4444444444 Quote rule 0
=====
Sampled 10049 rows (handled \n inside quoted fields) at 101 jump points including middle and very end
Bytes from first data row on line 2 to the end of last row: 47000004016
Line length: mean=79727.22 sd=32260.00 min=12804 max=153029
Estimated nrow: 47000004016 / 79727.22 = 589511
Initial alloc = 1179022 rows (589511 + 100%) using bytes/max(mean-2*sd,min) clamped between [1.1*estn, 2.0*estn]
=====
Type codes (colClasses) : 444444444444444444444444444444444444444444444444444444444444444444444444444444444444444444...4444444444
Type codes (drop|select) : 444444444444444444444444444444444444444444444444444444444444444444444444444444444444444444...4444444444
Allocating 12785 column slots (12785 - 0 dropped)
Reading 432 chunks of 103.756MB (1364 rows) using 144 threads
Read 872505 rows x 12785 columns from 43.772GB file in 02:17.726 wall clock time (can be slowed down by any other open apps even if seemingly idle)
Final type counts
0 : drop
0 : logical
0 : integer
0 : integer64
12785 : double
0 : character
Thread buffers were grown 67 times (if all 144 threads each grew once, this figure would be 144)
=============================
0.000s ( 0%) Memory map
0.099s ( 0%) sep, ncol and header detection
11.057s ( 8%) Column type detection using 10049 sample rows
0.899s ( 1%) Allocation of 872505 rows x 12785 cols (112.309GB) plus 37.433GB of temporary buffers
125.671s ( 91%) Reading data
0.000s ( 0%) Rereading 0 columns due to out-of-sample type exceptions
137.726s Total
Test 2:
Now using colClasses=list("numeric"=1:12785)
Timing was improved by few seconds...
DT<-fread('dt.daily.4km.csv', colClasses=list("numeric"=1:12785), verbose=TRUE)
Allocating 12785 column slots (12785 - 0 dropped)
Reading 432 chunks of 103.756MB (1364 rows) using 144 threads
Read 872505 rows x 12785 columns from 43.772GB file in 01:43.028 wall clock time (can be slowed down by any other open apps even if seemingly idle)
Final type counts
0 : drop
0 : logical
0 : integer
0 : integer64
12785 : double
0 : character
Thread buffers were grown 67 times (if all 144 threads each grew once, this figure would be 144)
=============================
0.000s ( 0%) Memory map
0.092s ( 0%) sep, ncol and header detection
11.009s ( 11%) Column type detection using 10049 sample rows
0.332s ( 0%) Allocation of 872505 rows x 12785 cols (112.309GB) plus 37.433GB of temporary buffers
91.595s ( 89%) Reading data
0.000s ( 0%) Rereading 0 columns due to out-of-sample type exceptions
103.028s Total
Ok great - we're getting there. The corrected sample size plus increasing it to 100 lines at 100 points (10,000 sample lines) was enough to guess the types correctly then - great. Since there are 12,875 columns and the line length is average 80,000 characters in the 44GB file, it it took 11s to sample. But that time was worth it because it avoided a reread which would have taken an extra 90s. We'll stick with that then.
I'm thinking the 2nd time is faster just because it was the 2nd time and your operating system has warmed up and cached the file. Anything else at all running on the box will affect the wall clock timings. That's addressed by running 3 identical consecutive runs of the 1st test. Then changing one thing only and running 3 identical consecutive runs again. At 44GB size you'll see a lot of natural variance. 3 runs is usually enough to make a conclusion but it can be a black art.
Yes the 112GB in memory vs 44GB in disk is partly because the data is bigger in memory because all the columns are type double; there is no in-memory compression in R and there are quite a lot of NA values which take no space in this CSV (just ",,") but 8 bytes in memory. However, it should be 83GB not 112GB (872505 rows x 12785 cols x 8 byte doubles / 1024^3 = 83GB). That 112GB is what was allocated based on the mean and standard deviation of the line lengths. Based on the average line length, it estimated it would be 589,511 rows which would have been too low. The line length variance was so high that the clamp came into effect at +100%. 58,9511 * 2 = 1,179,022 * 12785 * 8 / 1024^3 = 112GB. In the end it found that 872,505 are in the file. But it isn't freeing up the free space. I'll fix that. (TODO1)
When you specified the column type for all the columns, it still sampled, though. It should skip sampling when user has specified every column. (TODO2)
Since all your data is double, it's making 11 billion calls to C library function strtod(). The long wished for specialization of that function should in theory make a significant speedup for this file. (DONE)
Thanks for that, great explanation. Let me know if you want me to test something else with this file. I'm working on another data set that is more on the long-format, with ~21 million rows x 1432 columns.
NAME NROW NCOL MB
[1,] DT 21,812,625 1,432 238,310
Adding an additional data point to this. 89G .tsv, peak memory usage during loading is ~180G. I think this is expected as there are many NA and double.
I am also happy to test on this.
Ubuntu 16.04 64bit / Linux 4.4.0-71-generic
R version 3.3.2 (2016-10-31)
data.table 1.10.5 IN DEVELOPMENT built 2017-04-04 14:27:46 UTC
Architecture: x86_64
CPU op-mode(s): 32-bit, 64-bit
Byte Order: Little Endian
CPU(s): 64
On-line CPU(s) list: 0-63
Thread(s) per core: 2
Core(s) per socket: 16
Socket(s): 2
NUMA node(s): 2
Vendor ID: GenuineIntel
CPU family: 6
Model: 79
Model name: Intel(R) Xeon(R) CPU E5-2686 v4 @ 2.30GHz
Stepping: 1
CPU MHz: 2699.984
CPU max MHz: 3000.0000
CPU min MHz: 1200.0000
BogoMIPS: 4660.70
Hypervisor vendor: Xen
Virtualization type: full
L1d cache: 32K
L1i cache: 32K
L2 cache: 256K
L3 cache: 46080K
NUMA node0 CPU(s): 0-15,32-47
NUMA node1 CPU(s): 16-31,48-63
Flags: fpu vme de pse tsc msr pae mce cx8 apic sep mtrr pge mca cmov pat pse36 clflush mmx fxsr sse sse2 ht syscall nx pdpe1gb rdtscp lm constant_tsc rep_good nopl xtopology nonstop_tsc aperfmperf eagerfpu pni pclmulqdq monitor est ssse3 fma cx16 pcid sse4_1 sse4_2 x2apic movbe popcnt tsc_deadline_timer aes xsave avx f16c rdrand hypervisor lahf_lm abm fsgsbase bmi1 hle avx2 smep bmi2 erms invpcid rtm xsaveopt ida
Parameter na.strings == <<NA>>
None of the 1 na.strings are numeric (such as '-9999').
Input contains no \n. Taking this to be a filename to open
File opened, filesize is 88.603947 GB.
Memory mapping ... ok
Detected eol as \n only (no \r afterwards), the UNIX and Mac standard.
Positioned on line 1 starting: <<allele prediction_uuid sample_>>
Detecting sep ...
sep=='\t' with 101 lines of 76 fields using quote rule 0
Detected 76 columns on line 1. This line is either column names or first data row (first 30 chars): <<allele prediction_uuid sample_>>
All the fields on line 1 are character fields. Treating as the column names.
Number of sampling jump points = 101 because 95137762779 bytes from row 1 to eof / (2 * 24414 jump0size) == 1948426
Type codes (jump 000) : 5555542444111145424441111444111111111111111111111111111111111111111111111111 Quote rule 0
Type codes (jump 009) : 5555542444114445424441144444111111111111111111111111111111111111111111111111 Quote rule 0
Type codes (jump 042) : 5555542444444445424444444444111111111111111111111111111111111111111111111111 Quote rule 0
Type codes (jump 048) : 5555544444444445444444444444225225522555545111111111111111111111111111111111 Quote rule 0
Type codes (jump 083) : 5555544444444445444444444444225225522555545254452454411154454452454411154455 Quote rule 0
Type codes (jump 085) : 5555544444444445444444444444225225522555545254452454454454454452454454454455 Quote rule 0
Type codes (jump 100) : 5555544444444445444444444444225225522555545254452454454454454452454454454455 Quote rule 0
=====
Sampled 10028 rows (handled \n inside quoted fields) at 101 jump points including middle and very end
Bytes from first data row on line 2 to the end of last row: 95137762779
Line length: mean=465.06 sd=250.27 min=198 max=929
Estimated nrow: 95137762779 / 465.06 = 204571280
Initial alloc = 409142560 rows (204571280 + 100%) using bytes/max(mean-2*sd,min) clamped between [1.1*estn, 2.0*estn]
=====
Type codes (colClasses) : 5555544444444445444444444444225225522555545254452454454454454452454454454455
Type codes (drop|select) : 5555544444444445444444444444225225522555545254452454454454454452454454454455
Allocating 76 column slots (76 - 0 dropped)
Reading 90752 chunks of 1.000MB (2254 rows) using 64 threads
If it helps, here are results on a very long database: 419,124,196 x 42 (~2^34) with one header row and colClasses passed.
> library(data.table)
data.table 1.10.5 IN DEVELOPMENT built 2017-09-27 17:12:56 UTC; travis
The fastest way to learn (by data.table authors): https://www.datacamp.com/courses/data-analysis-the-data-table-way
Documentation: ?data.table, example(data.table) and browseVignettes("data.table")
Release notes, videos and slides: http://r-datatable.com
> CC <- c(rep('integer', 2), rep('character', 3),
+ rep('numeric', 2), rep('integer', 3),
+ rep('character', 2), 'integer', 'character', 'integer',
+ rep('character', 4), rep('numeric', 11), 'character',
+ 'numeric', 'character', rep('numeric', 2),
+ rep('integer', 3), rep('numeric', 2), 'integer',
+ 'numeric')
> P <- fread('XXXX.csv', colClasses = CC, header = TRUE, verbose = TRUE)
Input contains no \n. Taking this to be a filename to open
[01] Check arguments
Using 40 threads (omp_get_max_threads()=40, nth=40)
NAstrings = [<<NA>>]
None of the NAstrings look like numbers.
show progress = 1
0/1 column will be read as boolean
[02] Opening the file
Opening file XXXXcsv
File opened, size = 51.71GB (55521868868 bytes).
Memory mapping ... ok
[03] Detect and skip BOM
[04] Arrange mmap to be \0 terminated
\r-only line endings are not allowed because \n is found in the data
[05] Skipping initial rows if needed
Positioned on line 1 starting: <<X,X,X,X>>
[06] Detect separator, quoting rule, and ncolumns
Detecting sep ...
sep=',' with 100 lines of 42 fields using quote rule 0
Detected 42 columns on line 1. This line is either column names or first data row. Line starts as: <<X,X,X,X>>
Quote rule picked = 0
fill=false and the most number of columns found is 42
[07] Detect column types, good nrow estimate and whether first row is column names
'header' changed by user from 'auto' to true
Number of sampling jump points = 101 because (55521868866 bytes from row 1 to eof) / (2 * 13006 jump0size) == 2134471
Type codes (jump 000) : 5161010775551055105101111111111111110110771117717 Quote rule 0
Type codes (jump 022) : 5561010775551055105101111111111111110110771117717 Quote rule 0
Type codes (jump 030) : 5561010775551055105101010107517171151110110771117717 Quote rule 0
Type codes (jump 037) : 5561010775551055105101010107517771171110110771117717 Quote rule 0
Type codes (jump 073) : 5561010775551055105101010107517771177110110771117717 Quote rule 0
Type codes (jump 093) : 5561010775551055105101010107717771177110110771117717 Quote rule 0
Type codes (jump 100) : 5561010775551055105101010107717771177110110771117717 Quote rule 0
=====
Sampled 10049 rows (handled \n inside quoted fields) at 101 jump points
Bytes from first data row on line 1 to the end of last row: 55521868866
Line length: mean=132.68 sd=6.00 min=118 max=425
Estimated number of rows: 55521868866 / 132.68 = 418453923
Initial alloc = 460299315 rows (418453923 + 9%) using bytes/max(mean-2*sd,min) clamped between [1.1*estn, 2.0*estn]
=====
[08] Assign column names
[09] Apply user overrides on column types
After 11 type and 0 drop user overrides : 551010107755510105105101010107777777777710710775557757
[10] Allocate memory for the datatable
Allocating 42 column slots (42 - 0 dropped) with 460299315 rows
[11] Read the data
jumps=[0..52960), chunk_size=1048373, total_size=55521868441
Read 98%. ETA 00:00
[12] Finalizing the datatable
Read 419124195 rows x 42 columns from 51.71GB (55521868868 bytes) file in 13:42.935 wall clock time
Thread buffers were grown 0 times (if all 40 threads each grew once, this figure would be 40)
Final type counts
0 : drop
0 : bool8
0 : bool8
0 : bool8
0 : bool8
11 : int32
0 : int64
19 : float64
0 : float64
0 : float64
12 : string
=============================
0.000s ( 0%) Memory map 51.709GB file
0.016s ( 0%) sep=',' ncol=42 and header detection
0.016s ( 0%) Column type detection using 10049 sample rows
188.153s ( 23%) Allocation of 419124195 rows x 42 cols (125.177GB)
634.751s ( 77%) Reading 52960 chunks of 1.000MB (7901 rows) using 40 threads
= 0.121s ( 0%) Finding first non-embedded \n after each jump
+ 17.036s ( 2%) Parse to row-major thread buffers
+ 616.184s ( 75%) Transpose
+ 1.410s ( 0%) Waiting
0.000s ( 0%) Rereading 0 columns due to out-of-sample type exceptions
822.935s Total
> memory.size()
[1] 134270.3
> rm(P)
> gc()
used (Mb) gc trigger (Mb) max used (Mb)
Ncells 585532 31.3 5489235 293.2 6461124 345.1
Vcells 1508139082 11506.2 20046000758 152938.9 25028331901 190951.1
> memory.size()
[1] 87.56
> P <- fread('XXXX.csv', colClasses = CC, header = TRUE, verbose = TRUE)
Input contains no \n. Taking this to be a filename to open
[01] Check arguments
Using 40 threads (omp_get_max_threads()=40, nth=40)
NAstrings = [<<NA>>]
None of the NAstrings look like numbers.
show progress = 1
0/1 column will be read as boolean
[02] Opening the file
Opening file XXXX.csv
File opened, size = 51.71GB (55521868868 bytes).
Memory mapping ... ok
[03] Detect and skip BOM
[04] Arrange mmap to be \0 terminated
\r-only line endings are not allowed because \n is found in the data
[05] Skipping initial rows if needed
Positioned on line 1 starting: <<X,X,X,X>>
[06] Detect separator, quoting rule, and ncolumns
Detecting sep ...
sep=',' with 100 lines of 42 fields using quote rule 0
Detected 42 columns on line 1. This line is either column names or first data row. Line starts as: <<X,X,X,X>>
Quote rule picked = 0
fill=false and the most number of columns found is 42
[07] Detect column types, good nrow estimate and whether first row is column names
'header' changed by user from 'auto' to true
Number of sampling jump points = 101 because (55521868866 bytes from row 1 to eof) / (2 * 13006 jump0size) == 2134471
Type codes (jump 000) : 5161010775551055105101111111111111110110771117717 Quote rule 0
Type codes (jump 022) : 5561010775551055105101111111111111110110771117717 Quote rule 0
Type codes (jump 030) : 5561010775551055105101010107517171151110110771117717 Quote rule 0
Type codes (jump 037) : 5561010775551055105101010107517771171110110771117717 Quote rule 0
Type codes (jump 073) : 5561010775551055105101010107517771177110110771117717 Quote rule 0
Type codes (jump 093) : 5561010775551055105101010107717771177110110771117717 Quote rule 0
Type codes (jump 100) : 5561010775551055105101010107717771177110110771117717 Quote rule 0
=====
Sampled 10049 rows (handled \n inside quoted fields) at 101 jump points
Bytes from first data row on line 1 to the end of last row: 55521868866
Line length: mean=132.68 sd=6.00 min=118 max=425
Estimated number of rows: 55521868866 / 132.68 = 418453923
Initial alloc = 460299315 rows (418453923 + 9%) using bytes/max(mean-2*sd,min) clamped between [1.1*estn, 2.0*estn]
=====
[08] Assign column names
[09] Apply user overrides on column types
After 11 type and 0 drop user overrides : 551010107755510105105101010107777777777710710775557757
[10] Allocate memory for the datatable
Allocating 42 column slots (42 - 0 dropped) with 460299315 rows
[11] Read the data
jumps=[0..52960), chunk_size=1048373, total_size=55521868441
Read 98%. ETA 00:00
[12] Finalizing the datatable
Read 419124195 rows x 42 columns from 51.71GB (55521868868 bytes) file in 05:04.910 wall clock time
Thread buffers were grown 0 times (if all 40 threads each grew once, this figure would be 40)
Final type counts
0 : drop
0 : bool8
0 : bool8
0 : bool8
0 : bool8
11 : int32
0 : int64
19 : float64
0 : float64
0 : float64
12 : string
=============================
0.000s ( 0%) Memory map 51.709GB file
0.031s ( 0%) sep=',' ncol=42 and header detection
0.000s ( 0%) Column type detection using 10049 sample rows
28.437s ( 9%) Allocation of 419124195 rows x 42 cols (125.177GB)
276.442s ( 91%) Reading 52960 chunks of 1.000MB (7901 rows) using 40 threads
= 0.017s ( 0%) Finding first non-embedded \n after each jump
+ 12.941s ( 4%) Parse to row-major thread buffers
+ 262.989s ( 86%) Transpose
+ 0.495s ( 0%) Waiting
0.000s ( 0%) Rereading 0 columns due to out-of-sample type exceptions
304.910s Total
> memory.size()
[1] 157049.7
> sessionInfo()
R version 3.4.2 beta (2017-09-17 r73296)
Platform: x86_64-w64-mingw32/x64 (64-bit)
Running under: Windows Server >= 2012 x64 (build 9200)
Matrix products: default
locale:
[1] LC_COLLATE=English_United States.1252 LC_CTYPE=English_United States.1252
[3] LC_MONETARY=English_United States.1252 LC_NUMERIC=C
[5] LC_TIME=English_United States.1252
attached base packages:
[1] stats graphics grDevices utils datasets methods base
other attached packages:
[1] data.table_1.10.5
loaded via a namespace (and not attached):
[1] compiler_3.4.2 tools_3.4.2
A couple of notes. I would have suggested putting [09] before [07] in that if colClasses are passed there isn't a reason to check. Also, Windows showed about 160GB in use after each run. memory.size() probably does some cleaning. With 532GB RAM on this server, memory caching may have what to do with the increase in speed on the second run. Hope that helps.
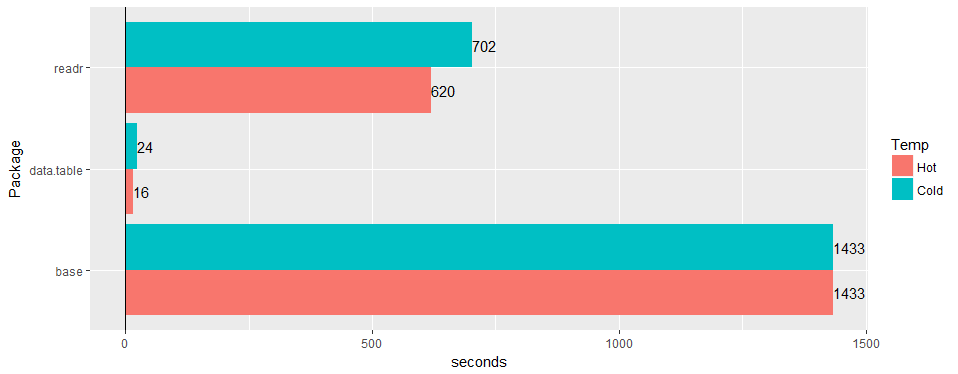
Test on a wide "real-world" table (hospital data): 30M rows × 125 columns v readr ‘1.2.0’ and read.csv 3.4.3.
any chance to confirm issue is still valid on 1.11.4? or code to produce example data.
This one is all resolved as far as I know. @geponce please update if not.
TODO1 above now filed as #3024
TODO2 above now filed as #3025
Most helpful comment
Test on a wide "real-world" table (hospital data): 30M rows × 125 columns v readr ‘1.2.0’ and
read.csv3.4.3.